Research

One of the main features of eukaryotic cells is the compartmentation between nucleus and cytoplasm. In this way the nuclear intron containing pre-mRNAs are not translated. Quality control mechanisms ensure that pre-mRNAs remain in the nucleus until splicing and processing is completed. However, in case intron containing mRNAs reach the cytoplasm, this could be harmful for cells and in higher eukaryotes this could lead to diseases such as cancer and neurodegenerative diseases.
In our work we focus on mRNA maturation the uncover the underlying quality control mechanisms in the eukaryotic model organism Saccharomyces cerevisiae. Quality control exists in the nucleus and involves the guard proteins and also in the cytoplasm, in which the ribosome is involved, called NoGo- NoStop- and NonsenseMediated-decay. Additional topics are analyses of several non-coding RNAs and their functions, including the antisense RNAs, the RNA of the telomease, snRNAs and snoRNAs. We use classical genetic and molecular biology methods in combination with cell biology and biochemical methods. A major goal is the identification of novel proteins acting in these processes and another important aspect is the functional characterization of the known factors, including the investigation of the underlying mechanisms.
Research focus topics:
•Identification and characterization of RNA-quality control factors
•Analyses of RNA transport pathways
•Mechanistic analyses of RNA quality control
•No go-, no stop- and nonsense mediated decay
•non-coding (nc)RNAs
How do mRNAs get out of the nucleus?
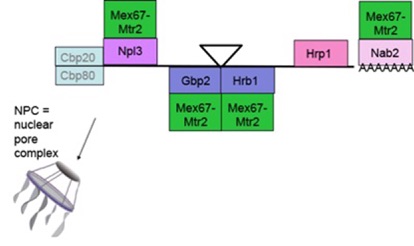
The compartmentalization of eukaryotic cells into nucleus and cytoplasm necessitates the movement of proteins and RNAs across the nuclear envelope. It has to be assured that intron containing pre messenger RNAs are retained in the nucleus until processing is completed. Only fully processed and spliced mRNAs should be transported into the cytoplasm and translated at the ribosomes. The otherwise resulting gene products can be toxic to cells and harmful to the organism. Several examples exist where not fully processed pre-mRNAs reach the cytoplasm, resulting in diseases like cancer or neurodegenerative diseases. One of our projects aims to identify and characterize the export-competent messenger ribonucleoprotein complexes (mRNPs) that are transported into the cytoplasm. Known yeast shuttling mRNA binding proteins include the cap binding complex (CBC), Hrp1, Nab2 and the three serine/arginine (SR)-type mRNA binding proteins Npl3, Gbp2 and Hrb1. The export receptor heterodimer Mex67-Mtr2 is recruited to the RNA by the export adapter proteins and docks the ribonucleoparticle to the nuclear pore complex (NPC).
Which other mRNA binding proteins are involved in the mRNA export?
We use the model organism Saccharomyces cerevisiae to identify novel transport factors by performing several genetic screens and have already identified several interesting candidate genes which are currently under investigation.
What are the functions of each individual mRNA binding protein in this process?
We have identified the SR-type shuttling mRNA binding proteins Gbp2 and Hrb1, that like Npl3 accompany the mRNA to the cytoplasm. Npl3 is recruited early to the pre-mRNA and associates upon maturation of the mRNA with the export receptor heterodimer Mex67-Mtr2 to mediate the passage through the NPC. Interestingly, although the export receptor is displaced by the helicase Dbp5 upon transit, Npl3 remains associated with the cytoplasmic mRNA and supports the formation of the ribosomes on the AUG start codons of the mRNAs. This involved its dimerization. Open questions are how and when does Npl3 recruit Mex67 and how does it later on mediate monosome formation? For this we investigate modifications of the proteins and functional studies in mutant backgrounds. Moreover, we found that Npl3 not only transports mRNAs, but it also exports the 60S pre-ribosomal subunits to the cytoplasm. The interconnection of these cellular events are part of our ongoing research.
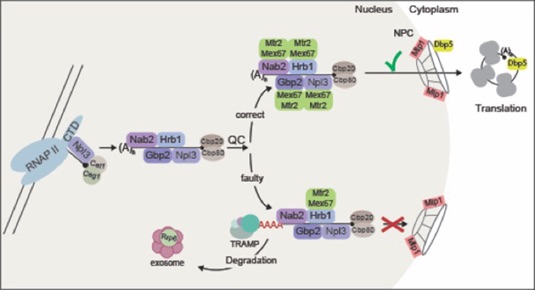
How does nuclear quality control of mRNAs work?
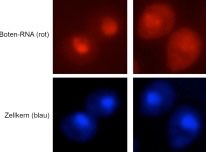
In contrast to Npl3, which is recruited to the mRNA via RNA-polymerase II, the other two SR-proteins Gbp2 and Hrb1 are recruited to the pre-mRNA via the THO complex, involved in transcription. We could recently show that their RNA binding is stabilized on pre-mRNAs that are spliced and that their binding is required for quality control of these RNA molecules. In fact, when Gbp2 and Hrb1 are absent, we found that intron containing false RNAs are exported into the cytoplasm. Both factors recruit the degradation machinery (TRAMP-complex and the nuclear exosome) to false mRNAs. On correct mRNAs the guard-proteins Gbp2 and Hrb1 instead recruit the export receptor Mex67 to promote transport of the mature mRNAs into the cytoplasm. The exact mechanisms are currently unclear and one topic in our lab.
Please visit our press release.
What happens during stress?
While cells grow well in a narrow range of physiological conditions, surviving extreme conditions is a challenge. Stressed cells express instantly chaperones that stabilize proteins and help to overcome this devastating condition. To ensure the preferential synthesis of such heat-shock (HS) proteins, cells inhibit transcription, pre-mRNA processing and nuclear export of non-HS transcripts, while stress-specific mRNAs are exclusively exported and translated. Surprisingly, it was largely unknown how cells manage the selective retention of regular transcripts and the simultaneous rapid export of HS mRNAs. We have shown that cellular stress induces the dissociation of Mex67 and its adapter proteins from regular mRNAs to prevent general mRNA export. At the same time, HS mRNAs are rapidly exported in association with Mex67, without the need for adapters. The immediate co-transcriptional loading of Mex67 onto HS mRNAs involves Hsf1 (heat shock transcription factor), which binds to heat shock promoter elements (HSEs) in stress-responsive genes. Importantly, the difference between both export modes is that adapter protein-bound mRNAs are quality controlled, while stress-specific transcripts are not. In fact, regular mRNAs are converted into uncontrolled stress-responsive transcripts by expression from a HS promoter, suggesting that the fate of being quality controlled is encrypted therein. Under normal conditions, Mex67-adapter proteins are recruited for RNA surveillance, allowing only quality-controlled mRNAs to associate with Mex67 and leave the nucleus. Thus, at the cost of an error-free mRNA formation, HS mRNAs are exported and translated without delay, allowing cells to survive extreme situations.
False mRNAs are retained in the nucleus of wild type cells (left) but leak into the cytoplasm in cells lacking Gbp2 and Hrb1 (right).
Please visit our press release
What is the function of the shuttling mRNA binding proteins during translation?
With our recent studies we gained insights into the transformation of an exporting to a translating mRNA/protein complexes: Upon arrival in the cytoplasm bulk of the shuttling mRNA binding proteins leave the mRNA, whereas approximately 30% of the extracted protein of the three SR-proteins Npl3, Gbp2 and Hrb1 remain bound to the mRNA during translation. Moreover, we found that the DEAD-box RNA-helicase Dbp5 is associated with polyribosomes.
In studies with the DEAD-box RNA-helicase Dbp5 we have recently uncovered a novel function for Dbp5 in translation. Besides its well established function in mRNA export we have identified Dbp5 as a novel player in translation termination. Dbp5 interacts genetically with both release factors eRF1 and eRF3 and Pab1. A physical interaction was specifically detected with eRF1. We have shown that during translation termination the helicase activity of Dbp5 is required for efficient stop-codon recognition, and intact Dbp5 is essential for the recruitment of eRF3 into termination complexes. Therefore, Dbp5 controls the eRF3-eRF1 interaction and thus eRF3-mediated downstream events.
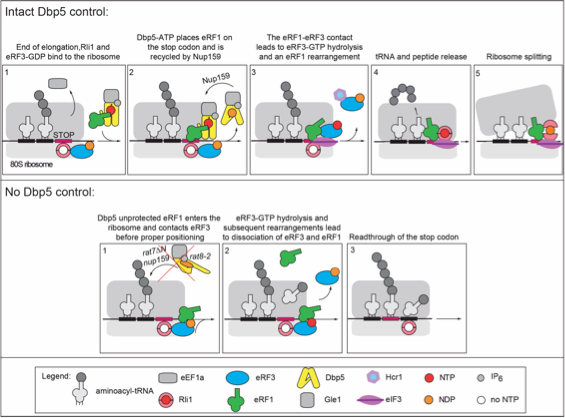
Please visit our press release.
Another unexpected protein that we have shown acts in translation termination is the iron-sulfur cluster protein Rli1. In contrast to Dbp5, Rli1 interacts with both eRF1 and eRF3 to promote termination. Thus, it becomes clear that the process of translation termination is more complex than expected and needs further investigations.
With this project, we are part of the DFG collaborative research center SFB 860 (Integrative structural biology of dynamic macromolecular assemblies)
How are non-coding RNAs generated and what is their function?
Besides the protein encoding messenger RNAs many non-coding RNAs exist in cells that have a many regulatory functions. Best known examples are e.g. the snRNAs that are involved in splicing or the telomerase RNA TLC1 that is important for Telomere maintenance but many other non-coding RNAs exist. We are studying how they are synthesized, processed and assembled with proteins to functional units.
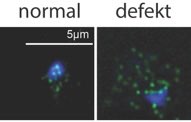
Most recently we found that TLC1 shuttles to the cytoplasm via mRNA export factors where it associates with the Est-proteins that are important for the functionality of the telomerase and export mutants have shortened telomeres.
Localization of a protein of the telomerase (Est1, green) in a wild type cell (normal) and an export mutant (defect). The DNA of the nucleus is shown in blue.
Please visit our press release
Please visit also: https://www.uni-goettingen.de/de/3240.html?id=5481
Selected publications:
Grosse, S., Lu, Y-Y., Coban, I., Neumann, B. and Krebber, H. (2020) Nuclear SR-protein mediated mRNA quality control is continued in the cytoplasmic nonsense-mediated decay. RNA-Biology, Jan 7; 1-18 doi: 10.1080/15476286.2020.1851506
Beißel, C., Grosse S. and Krebber H. (2020) Dbp5/DDX19 between translational readthrough and nonsense mediated decay. Int. J. Mol. Sci. , 21, 1085; doi:10.3390/ijms21031085
Frumkin I, Yofe I, Bar-Ziv R, Gurvich Y, Lu YY, Voichek Y, Towers R, Schirman D, Krebber H, Pilpel Y. (2019) Evolution of intron splicing towards optimized gene expression is based on various Cis- and Trans-molecular mechanisms. PLoS Biol. Aug 23;17(8):e3000423. doi: 10.1371/journal.pbio.3000423.
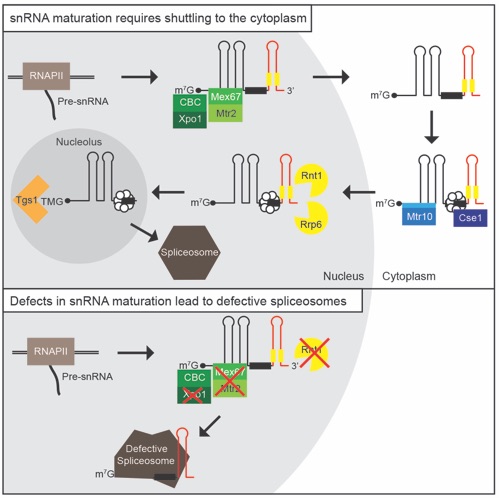
Becker, D., Hirsch, A.G., Bender, L., Lingner, T., Salinas, G. and Krebber H., (2019) Nuclear pre-snRNA export is an essential quality assurance mechanism for functional spliceosomes. Cell Reports,
https://doi.org/10.1016/j.celrep.2019.05.03
Beissel, C., Neumann, B., Uhse, S., Hampe, I., Karki, P. and Krebber, H. (2019) Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3. Nucleic Acids Research. https://doi: 10.1093/nar/gkz177
Zander, G. and Krebber, H. (2017) Quick or Quality? How mRNAs escapes nuclear quality control during stress. RNA Biology, Jul 14:1-7.
Zander, G., Hackmann, A., Bender, L., Becker, D., Lingner, T., Salinas, G. and Krebber, H. (2016) mRNA quality control is bypassed for an immediate export of stress responsive transcripts. Nature, 540: 593-596.
Wu, H., Becker, D. and Krebber H. (2014) Telomerase RNA TLC1 shuttling to the cytoplasm requires mRNA export factors and is important for telomere maintenance. Cell Reports 8: 1-9.
Hackmann A, Wu H, Schneider UM, Meyer K, Jung K and Krebber H. (2014) Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nature Communications 5:3123.
Baierlein C, Hackmann A, Gross T, Henker L, Hinz F and Krebber H. (2013) Monosome formation during translation initiation requires the serine/arginine-rich protein Npl3. Mol Cell Biol. 33(24):4811-23.
Tieg, B. and Krebber, H. (2013) Dbp5 - From nuclear export to translation. Biochem. Biophys. Acta 1829(8):791-798.
Hackmann, A., Gross, T., Baierlein, C. and Krebber, H. (2011) The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO-Reports, 12(10): 1024-1031.
Baierlein, C. and Krebber, H. (2010) Translation termination: New factors and insights. RNA-Biology 7:issue 5: 548 - 550.
Khoshnevis, S., Gross, T., Rotte, C., Baierlein, C., Ficner, R., Krebber, H. (2010) The iron-sulphur protein RNase L inhibitor functions in translation termination EMBO Reports, 11(3): 214-219.
Gross, T., Siepmann, A., Sturm, D., Windgassen, M., Scarelli, J., Cole C.N., Seedorf, M., und Krebber, H. (2007) The DEAD-box RNA-helicase Dbp5 functions in translation termination. Science, 315: 646-649.
Windgassen, M., Sturm, D., Cajigas, I.J., González, C.I, Seedorf, M., Bastians, H. und Krebber, H. (2004) Yeast shuttling SR-proteins Npl3p, Gbp2p and Hrb1p are part of the translated mRNAs and Npl3p can function as a translational repressor. Mol. Cell. Biol., 24 (23): 10479-10491.
Hacker, S., and Krebber, H., (2004) Differential export requirements for shuttling SR-type mRNA binding proteins. J. Biol. Chem. , 279 (7):5049-5052.
Windgassen M. and Krebber, H. (2003) Identification of Gbp2p as a novel poly(A)+RNA binding protein in yeast involved in the cytoplasmic delivery of mRNAs. EMBO Reports, 4 (3): 278-283.
Heike Krebber on
ResearchGate : https://www.researchgate.net/profile/Heike_Krebber